

KEVIN DUERINCK SURNAME
DNA PROJECT RESULTS

Welcome to the DUERINCK SURNAME DNA PROJECT RESULTS (project started year 2000)
[LAST REVISED September 5, 2010. If a Duerinck or variant surname, join this project at: Duerinck Surname y-DNA Project Registration at FTDNA
Values for all DYS 464 alleles have been reduced by "1" per Family Tree DNA. List of y-DNA Surname Projects
NOTE: 25 marker (12 + 13) test taken by 5 participants, results of the extra 13 markers are listed in the second table. 37 markers for 1 participant, kit 281 (see third table), and 66 or 67 markers done in Phase IV for kit 281.
|
|
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
|
|
Kit |
Surname |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
Country |
|
281 |
Duerinck |
13 |
23 |
13 |
11 |
11 |
15 |
12 |
12 |
12 |
14 |
13 |
31 |
USA/Belgium |
|
282 |
Durinck |
13 |
24 |
13 |
11 |
11 |
15 |
12 |
12 |
12 |
14 |
13 |
31 |
Belgium |
|
285 |
Durinck |
13 |
24 |
13 |
11 |
11 |
15 |
12 |
12 |
12 |
14 |
13 |
31 |
Australia/Belgium |
|
1069 |
Dierick |
13 |
24 |
14 |
11 |
11 |
15 |
12 |
12 |
12 |
14 |
13 |
31 |
Belgium |
|
1070 |
Dierick |
13 |
24 |
13 |
11 |
11 |
15 |
12 |
12 |
12 |
14 |
13 |
31 |
Belgium |
|
288 |
Duerinck |
13 |
24 |
13 |
10 |
11 |
15 |
12 |
12 |
12 |
15 |
13 |
32 |
Belgium |
|
298 |
Dueringer |
13 |
24 |
13 |
10 |
11 |
14 |
12 |
12 |
12 |
14 |
13 |
30 |
USA |
|
297 |
Dieringer |
13 |
23 |
13 |
11 |
11 |
14 |
12 |
12 |
11 |
13 |
13 |
29 |
USA |
|
299 |
VonDuering |
13 |
23 |
13 |
10 |
11 |
14 |
12 |
12 |
11 |
13 |
13 |
28 |
USA/Germany |
|
337 |
Duering |
13 |
24 |
13 |
11 |
11 |
15 |
12 |
12 |
11 |
12 |
13 |
28 |
Germany |
|
283 |
Duerinckx |
12 |
23 |
13 |
10 |
13 |
16 |
11 |
14 |
11 |
13 |
11 |
29 |
Belgium |
|
287 |
Doehring |
13 |
22 |
14 |
10 |
16 |
16 |
11 |
13 |
11 |
13 |
12 |
29 |
USA |
|
|
|
13 |
14 |
15 |
16 |
17 |
18 |
19 |
20 |
21 |
22 |
23 |
24 |
25 |
|
|
Kit |
Surname |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
Country |
|
281 |
Duerinck |
17 |
9 |
10 |
11 |
11 |
25 |
15 |
19 |
30 |
15 |
16 |
17 |
18 |
USA/Belgium |
|
1069 |
Dierick |
16 |
9 |
10 |
11 |
11 |
25 |
15 |
19 |
30 |
15 |
15 |
17 |
18 |
Belgium |
|
285 |
Durinck |
17 |
9 |
10 |
11 |
11 |
25 |
15 |
18 |
31 |
15 |
15 |
17 |
18 |
Belgium/Australia |
|
288 |
Duerinck |
17 |
9 |
10 |
11 |
11 |
25 |
15 |
19 |
30 |
15 |
15 |
17 |
18 |
Belgium |
|
283 |
Duerinckx |
18 |
9 |
9 |
11 |
12 |
25 |
15 |
19 |
30 |
12 |
14 |
15 |
15 |
Belgium |
|
|
|
26 |
27 |
28 |
29 |
30 |
31 |
32 |
33 |
34 |
35 |
36 |
37 |
|
|
Kit |
Surname |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
Country |
|
281 |
Duerinck |
11 |
11 |
19 |
23 |
16 |
14 |
19 |
16 |
36 |
37 |
12 |
12 |
USA/Belgium |
|
1069 |
Dierick |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Belgium |
|
285 |
Durinck |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Belgium/Australia |
|
288 |
Duerinck |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Belgium |
|
283 |
Duerinckx |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
Belgium |
|
|
|
38 |
39 |
40 |
41 |
42 |
43 |
44 |
45 |
46 |
47 |
48 |
49 |
|
|
Kit |
Surname |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
Country |
|
281 |
Duerinck |
11 |
9 |
15 |
16 |
8 |
10 |
10 |
8 |
10 |
10 |
12 |
22 |
USA/Belgium |
|
|
|
50 |
51 |
52 |
53 |
54 |
55 |
56 |
57 |
58 |
59 |
60 |
61 |
|
|
Kit |
Surname |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
Country |
|
281 |
Duerinck |
25 |
16 |
10 |
12 |
12 |
15 |
8 |
12 |
22 |
20 |
14 |
12 |
USA/Belgium |
|
|
|
62 |
63 |
64 |
65 |
66 |
67 |
|
|
Kit |
Surname |
DYS |
DYS |
DYS |
DYS |
DYS |
DYS |
Country |
|
281 |
Duerinck |
12 |
14 |
11 |
11 |
12 |
12 |
USA/Belgium |
Phase IV for kit 281: Kit 281 results have now been posted, above.
Latest News: The Phase III results in the third table will be discussed at a later date. The first table shows that Dierick is related to Durinck (282, 285) and Duerinck (281). Based on the total of 25 markers, however, the relationships show a match of 21 out of 25 markers for kits 281, 1069, 285 and 288. Generally, we say there is relationship at 22 out of 25 markers. This is a new science, however, and we are all still learning, and we all basically have very close surnames. Let's wait and see what science brings in the near future to support any relatedness. We also encourage males in other clans to undergo testing. We do need representation from some of the Netherland clans as well.
* NOTE: DYS identification numbers for all twelve loci have been released and are shown, as well as the measured repeat values. You will notice that in the columns under each locus, the total number of "repeats" is shown. Thus, the numbers that you will see, the 13's, 23's, etcetera, are the repeats, or units of repeating DNA segments. For example, a repeat of 11 for loci DYS19 means that the sequence TAGA is repeated 11 times, which comes out to 190 bp (base pairs). The pound sign (#) by DYS19 means that the reported DYS394 is equivalent to DYS19 which I show here. Jobling, M.A. and Tyler-Smith, C. (1995) Trends Genet. 11, 449-456. Fathers and sons: the Y chromosome and human evolution.
[For information on DYS 464 a,b,c,d mutation analysis, see Georgia Kinney Bopp's page and L. David Roper's page]
One note regarding loci 389i and 389ii, reported by Doug Mumma: "the number of "repeats" reported as DYS389II includes the number of "repeats" reported at DYS389I. As a result, if a one-step mutation is observed at DYS389I, then DYS389II will also show a one-step mutation. The "double reporting" of the repeats observed at DYS389I is not desirable. To eliminate this "doubling", many researchers report DYS389II as either 389II-I or as 389AB, which then allows you to distinguish whether a mutation occurred on the 389I portion of the locus or on the other portion of the 389 locus. All you need to do is subtract the number of repeats reported for 389I from those reported for 389II and identify it as either 389AB or 389ii-i for clarity."
Let's put it this way. Below are 6 examples. For figuring out total mutations at each locus, you need to figure out mutations at 389i and 389ii. First, figure out the mutations for DYS389i by subtracting one kit's value from the other. Second, to see how many mutations are at 389ii, just subtract one kit's 389i value from the same kit's 389ii value, then do it with the other kit. Now compare. So here goes!:
Example 1 (389i and ii show values as greater than compared kit 281. These are the real
values on my site. The other examples below are fake) kit 281 (FTDNA) DYS389i DYS389ii Mutations 14 31 kit 288 (FTDNA) 15 32 1 Calculations: 14 vs. 15 is one mutation. In both cases 389ii - 389i = 17, so no mutation in 389ii. So total is 1 mutation. Example 2 (389i and ii show values as lesser than compared kit 281) kit 281 (FTDNA) DYS389i DYS389ii Mutations 14 31 kit 288 (FTDNA) 13 30 1 Calculations: same as above, only different direction; still one mutation. Example 3 (1 value lesser (389i), 1 value greater (389ii) than compared kit 281) kit 281 (FTDNA) DYS389i DYS389ii Mutations 14 31 kit 288 (FTDNA) 13 32 3 Calculations: There is one mutation in 389i (14 vs. 13), and there is an additional change(s) in 389ii. In this case, 31-14=17, vs. 32-13=19. So there are 2 changes in 389ii and 1 in 389i, so 3 mutations total. Example 4 (1 value lesser (389ii), 1 value greater (389i) than compared kit 281) kit 281 (FTDNA) DYS389i DYS389ii Mutations 14 31 kit 288 (FTDNA) 15 30 3 Calculations: Same as above, only different direction. One mutation in 389ii, and then for 389ii (31-14=17 vs. 30-15=15, so two mutations there). So 3 total. Example 5 (1 value greater (389i), 1 value equal (389ii) than compared kit 281--This
example was from the list awhile ago--2 mutations. Compare to example 6 below) kit 281 (FTDNA) DYS389i DYS389ii Mutations 14 31 kit 288 (FTDNA) 15 31 2 Calculations: One mutation at 389i, and for 389ii (31-14=17 vs. 31-15=16; i.e., 1 mutation), so 2 mutations total. Example 6 (1 value equal (389i), 1 value greater (389ii) than compared kit 281--The
"opposite" of example 5) kit 281 (FTDNA) DYS389i DYS389ii Mutations 14 31 kit 288 (FTDNA) 14 32 1 Calculations: One mutation. 31-14=17 vs. 32-14=18.
Let's give another example. Let's say there is a mutation at 389i but it is not showing up at 389ii. What does that mean? It means that there are two mutations in the DNA. You see, 389ii has two contributions that are added together. One of those is reported separately under 389i. If the second part happens to mutate in the opposite direction from the change in 389i, then the two mutations will appear to "cancel" in the sum reported as 389ii. By the way, thanks to Greg Bonner and FTDNA for their advice on these issues.
What does this all mean? Well, first, you must understand some nomenclature. For testing, we look at the y chromosome. We look at a locus, a location on the chromosome. At this locus, there might be 10 to 30 alleles, or identification markers. A male will have only one of these alleles, or markers (generally true, although there are instances where the male may have two marker segments (for example, DYS394) on their Y-chromosomes with different repeat values (Lindsay).
Second, you must understand that this project had a very small sample size of only 12 males (I add new data as new males join the Project--originally had 10 males tested). So where one of us may question the results as not being possible, we must remember the small sample size. Additional testing could put any questions to rest, especially if you test someone who is descended farther back from the same paternal line of ancestors. The Duerinckx and Doering surnames in this small sample are clearly not related to Duerinck and Durinck. Again, for Duerinckx I would recommend another male relation be tested who is descended from a common ancestor farther back in the paternal line.
The Duerinck surname from USA/Belgium and the Durinck surname from Belgium/Australia have a haplotype that is a 1 step difference (one mutation on one loci). What this means is that we are related. For an 11/12 match, there is a 50% probability that the most recent common ancestor (MRCA) lived no longer than 36 generations, which is also the MLE. This translates into 36 x 20 = 720 years. The range for an 11/12 match is 1 to 104 generations (85 generations at 90%, 104 at 95%). The question becomes, "who has the original haplotype?" One or both families would have to test a male who is descended from a male farther back in the paternal line in order to answer this question. As more markers are tested, let us say 20, the MRCA date range of 500 to 1500 years would be narrowed considerably. Based on new test results, the Dierick surname has a 1 step difference on 1 loci compared to Durinck. Compared to Duerinck (281), Dierick is a 1 step difference on 1 loci and a 1 step difference on another loci. I will say that Dierick is related to Durinck as well as Duerinck (281). It is also interesting that the Dierick surname contains the Atlantic Modal Haplotype. Along the Atlantic coast in Europe, the most common haplotype is
called 1.15, the 'Atlantic Modal Haplotype' or AMH, being Hg 1 (haplogroup 1, now known as R1b) with the following haplotype:
DYS 19 388 390 391 392 393
Repeats 14 12 24 11 13 13
The Duerinck surname from Belgium, another clan that had ancestors in the same town as mine around the 1600's, is a 3 step difference in their haplotype from my Duerinck haplotype. [When comparing one individual to another, if DYS389I and DYS389II differ by the same number of repeats, it should only be counted as one mutation (as in Duerinck 288). However, there can be a mutation at DYS389II that will not affect DYS389I]. Yet, Duerinck 288 is only a 2 step difference from the Durinck Belgium/Australia haplotype. So we had this family tested in Phase II for the full 25 markers.
The Dueringer and Dieringer haplotypes have 4 and 5 step differences from the Duerinck (281) haplotype and a 4 and 6 step difference from the Durinck haplotype.
Duering and Von Duering haplotypes are 7 steps away from the Duerinck (281) haplotype and 6 and 7 steps away from the Durinck haplotype.
The Duerinckx (test kit 283) and Doehring (test kit 287) haplotypes are off from the Durinck haplotype by 14 and 17 mutations, respectively. The number of mutations off of the Duerinck (281) haplotype would be similar.
As for Duerinckx, we have had a further test done. Duerinckx is a Med+ haplotype, meaning that their ancestors were of Mediterranean extraction, possibly either Jewish Cohanim, Levite or Arabic. Further testing showed that Duerinckx was 2 mutations off of the Cohen modal haplotype, indicating a Levite ancestry (per Bennett Greenspan). A Levite is a member of the tribe of Levi, not descended from Aaron. The Levite males were chosen to assist the Temple priests.
There is a lot of talk about haplotype versus haplogroup. Some definitions from Peter de Knijff's paper "Messages through Bottlenecks: On the Combined Use of Slow and Fast Evolving Polymorphic Markers on the Human Y Chromosome" [Am. J. Hum. Genetics, 67:1055-1061 (2000)]. "Distinct Y chromosomes identified by STR's are designated as 'haplotypes.'" Stated in another way: a haplotype is represented by the number of repeats at certain alleles, or markers on the y chromosome. "Distinct Y chromosomes, defined solely on the basis of (unique mutation events) UME character states, are designated as 'haplogroups.'" Each haplogroup consists of a variable number of Y chromosomes that share the same UME character state but vary in Y-STR haplotype. "Y chromosomes that are defined by the combination of UMEs [haplogroups]and combination of STRs [haplotypes] are called 'lineages.'"
Care must be taken when comparing results. There is always a possibility that someone may be comparing a haplotype of someone in a different haplogroup. In one-name studies this "threat" is diminished. However, if you are searching databases that have haplotypes, you have to be sure that you are comparing apples and apples, not apples and oranges--comparing those in your own haplogroup (which these databases do not yet provide for). While I thought that future testing should include more biallelic polymorphisms in order to determine haplogroup, Dr. Mark Jobling says what is needed are more microsatellites in testing, not biallelic polymorphisms. At present we are seeing people compare apples and oranges, saying that they may have a common ancestor with some other surname such as Duerinck, when in reality, we both may be in different haplogroups. I am currently reading 3 papers from the American Journal of Human Genetics on these issues: "Y-Chromosomal Diversity in Europe is Clinal and Influenced Primarily by Geography, Rather than by Language" by Rosser, et al. [AJHG vol 67 page 1526 (2000)]; "Estimating Scandinavian and Gaelic Ancestry in the Male Settlers" by Helgason et al. [AJHG vol 67 page 697 (2000)]; and "Messages through Bottlenecks: On the Combined Use of Slow and Fast Evolving Polymorphic Markers on the Human Y Chromosome" by de Knijff [AJHG vol 67 page 1055 (2000)].
In the Rosser study above, they looked at 3,616 Europeans in 47 populations, and found them to be in 12 Haplogroups: Hg1 (now R1b), Hg2, Hg3 (now R1a), Hg4, Hg7, Hg8, Hg9, Hg12, Hg16, Hg21, Hg22, and Hg26.
Out of 92 Belgians 63% were in Hg 1 (now R1b), 23% were in Hg 2, 4% in Hg 3 (now R1a), 5% in Hg 9, and there were traces of Hgs 21, 22 and 26.
Out of 84 Dutch 43% were Hg 1 (now R1b), 32% were Hg 2, 13% were Hg 3 (now R1a), 7% were Hg 9, 8% were Hg 21, and 1% Hg 22.
Out of 30 Germans (Bavaria was another category), 40% were Hg 1 (now R1b), 20% Hg 2, 30% Hg 3 (now R1a), 3% Hg 9, 3% Hg 16, and 3% Hg 26.
As Patrick Guinness of Ireland has noted, Hg 1 (now R1b) is known by other names, depending on which researcher you talk to:
Tatiana Karafet......1C, Am. Jour. Hum. Genet. 1999, vol. 64, p.822
Ornella Semino......Eu 18, Science 2000, vol. 290, p. 1155
Zoe Rosser............Hg 1, Am. J. Hum. Genet. 2000, vol. 67, p. 1526
Emmy Hill............Hg 1, Nature 2000, vol. 404, p. 351
It appears that HG 1 may be the main pre-Ice Age male element in western Europe (for y-chromosome analysis), similar to Prof. Sykes's Daughters of Eve (mtDNA analysis). HUGO, the gene nomenclature body, and the University College of London would like to resolve the differences in nomenclature for the haplogroups.
In order to understand MRCA, you must understand the following:
--MRCA (most recent common ancestor): the common ancestor between 2 people.
--generation: it means every 20 years here (lot of discussion on this point--most of us are inclined to believe that 25 years is a better estimate).
--Heyer principle: y chromosome mutations occur generally once every 500 generations per locus (or "per marker") (mutation rate of 0.2% or 0.002, Heyer et al. 1997)
--Example of Heyer principle: Using the Heyer principle, we would expect EACH locus to change 1 digit in 500 generations. For example, we would expect that a 5 (repeat units) in locus #1 would move to either a 4 or a 6 in that 9-10,000 year time frame (500 generations).
So, instead of ONE locus, how about looking at 12 loci (or 15 or 20 etc.)? If we use a 12 marker test, in 500 (generations) / 12 (markers) = 41.667 generations we would expect to have a change in 1 of the 12 markers by 1 digit. OR, to be in the "club" of Duerincks, if the Common Ancestor was farther back in time than 41.667 generations (a generation = 20 years), then it would be totally expected that a "real" related Duerinck (Kit 281 in table above) would have a 1 digit change in 835 years.
Getting back to the reasonableness of expecting a mutation to occur every 41 or so generations if a 12 marker test (testing 12 loci) is used, what does the mutation rate become if more markers are used? Simple. You must also remember that loci changes (mutations) can occur at any time. Using the y chromosome mutation rate PER MARKER of once every 500 generations, if 15 loci are measured, 500 divided by 15 = every 30 generations there could be a mutation at a loci. If 17 loci are measured, 500 divided by 17 = every 29 generations. At 20 loci being measured, 500 divided by 20 = every 25 generations there could be a mutation at a loci. Where 21 loci are measured and you have a 21/21 match, this indicates that there is a 50% probability that the MRCA is within 9 generations and a 90% probability that the MRCA is within 28 generations.
As genealogists we want to know when the MRCA between two people lived. We know that where all 12 markers match, there is a 50% probability that the MRCA was no longer than 14.5 generations (290 years), and a 90% probability that the MRCA was within the last 48 generations (63 generations at 95%). The range for a 12/12 match is 1 to 63 generations. Further, if a test uses 12 different loci (a 12 marker test), it is reasonable to expect a change to occur every 41 or so generations; however, these changes can take place at any time.
What if there is an 11/12 marker match? For an 11/12 match, there is a 50% probability that the MRCA lived no longer than 36 generations. This translates into 36 x 20 = 720 years. The range for an 11/12 match is 1 to 104 generations (85 generations at 90%, 104 at 95%).
What if there is a 10/12 match? For a 10/12 match, there is a 50% probability that the MRCA lived no longer than 61 generations. The range for a 10/12 match is 1 to 145 generations (122 at 90%, 145 at 95% probability).
We do know that if the results of 2 people are different by 1 mutation, that these people are related. If different by 2 mutations, probably related. Is there a difference if there is a 2 repeat value change on one loci OR if there is are 1 repeat value changes on two loci? As we see 3 or more mutations, the people who may be related are very distantly related. The concept of MRCA puts the relation back many generations, more like 1,000 to 2,000 years for 3 mutations. For 3 or more mutations out of a 12 marker test, people are not related from a genealogical standpoint.
Just an update when using 21 markers: if you match 21/21 then the likelihood of having a common ancestor with that person is that in 50% of the cases the common ancestor would have been within 8.3 generations. Of course it could be more recent and it could be farther back.
At a 25 marker match between 2 individual males (with the same or variant surname), there is a 50% probability that the most recent common ancestor is 7 generations back or less. We are speaking of confidence intervals. In other words, 50% will find their common ancestor within that range. If you want to be more confident (think of it as placing a gambling bet--when did the MRCA live?), the number of generations back to the MRCA would increase. How confident are you at 95% probability? 95% will find their common ancestor within 30 generations ago or less for a 25/25 marker match. You can find the MRCA within the range of 0 to 37 generations for a 25/25 marker match.
Did you ever wonder what DYS stands for? D = DNA, Y = Y-chromosome, S = (unique) segment [or "single copy sequence" according to "Forensic DNA Typing" by John Butler]. The DYS numbering scheme (e.g. DYS388, DYS390) for the Y-STR haplotype loci is controlled and administered by an international standards body called HUGO Human Gene Nomenclature Committee based at University College, London.
Here are most of the results represented in a phylogenetic tree. This graph was done by Ann Turner, GENEALOGY-DNA List Administrator, and is used here with her express permission. You will note the markers are DYS markers for the known markers and FT markers for those markers used by FTDNA that have not yet been made public. The software that was used is available at Fluxus-Engineering. The instruction page notes that in human Y STRs, compound STRs such as DYS389II should be resolved into its mutational subcomponents m, n, and q to avoid artefacts. In the graph below, the Reduced Median (RM) network method was used.
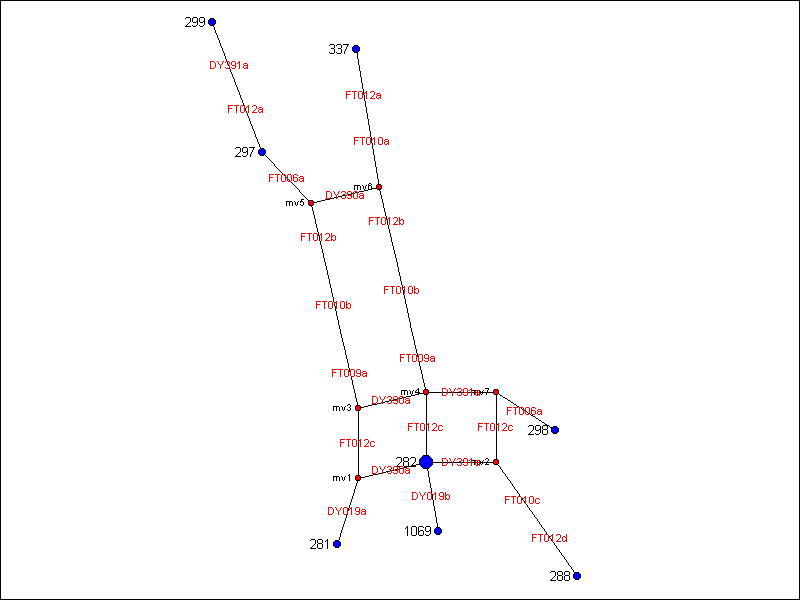
The above phylogenetic tree shows 2 clusters (tree needs to be updated showing closer relationships by eliminating an error in locus 19 which allele was previously 12 but has now been corrected to 13 in the results chart). Regarding the first cluster at the bottom, for Durinck/Duerinck/Dierick, the graph shows that these surnames originate from the family represented by participant number 282, Durinck. That includes numbers 281 Duerinck (me), 1069 Dierick and 288 Duerinck. It appears that participant 298 Dueringer, might be related to Durinck further back. I would caution the reader that under current technology, this last relationship may or may not be statistically significant--3 mutations away from Durinck--even though it is on the graph. More samples and more markers would have to be studied.
The second cluster, at the top of the graph, shows participants 299 Von Duering, 297 Dieringer, and 337 Duering to be more closely related than those participants in the first cluster and the bottom. Participants 283 and 287 were not graphed because they were the furthest away from any of the other participants and also because the software used started messing up.
Duerinck Surname y-DNA Project
Why DNA Testing For Genealogy and How To Manage It
Surname DNA Project Waiver/Release Form
State of Illinois Genetic Information Privacy Act -- Selected sections
Short Tandem Repeat (STR) Analysis
Medical Genetics and Genealogy: Genetic Diseases
Genetics and Human Migration Patterns
DNA Consulting (Panther-Yates)
Distribution of Blood Types (by Dennis O'Neil
Kerchner DNAPrint© Test Results Log Book
DNA Genealogy Timeline (from Georgia Bopp)
Chris Pomeroy's Chart of Company Markers Offered
Ron Lindsay's Chart of Company Markers Offered
Roper's in-depth review of BioGeographical Ancestry Testing
NIST's Short Tandem Repeat DNA Internet DataBase
YHRD - Y Chromosome Haplotype Reference Database (Supplants the YSTR databases--more robust search)
Leiden University's Regional Allele Frequencies Database
Custer's Y-STR Loci Allele Frequencies [spreadsheets, frequency graphs]
Free DNA matching Database [by the Genealogy Researcher Community]
SMGF Database Search [by the Sorenson Molecular Genealogy Foundation]
Time to MRCA by Family Tree DNA
Bruce Walsh's Time to Most Recent Common Ancestry Calculator
Blair DNA 101--see MRCA calculations
YBase Y-chromosome haplotype database
RootsWeb Genealogy-DNA ListServe [where we all hang out]
Brigham Young University DNA Project [current project will yield a huge database of results--whether anyone gets access to it, and how, remains to be seen]
Scylla's Genetic Links [includes databases, mapping, sequencing]
SNP Consortium [856,666 mapped SNPs; 607 are y-SNPs in 8th data release]
National Center for Biotechnology Information database [dbSNP--3 million SNPs]
"DNA for Family Historians", by Alan Savin Savin DNA book here
"Mapping Human History" by Steve Olson, May 2002.
"The Journey of Man: A Genetic Odyssey" by Dr. Spencer Wells
Cavalli-Sforza, L.L. and Cavalli-Sforza, F. The Great Human Diasporas: the History of Diversity and Evolution, Addison-Wesley Publishing Co., Reading, MA (1995).
"The Human Inheritance: Genes Language and Evolution" edited by Bryan Sykes. Published by Oxford University Press 1999
Sykes, Bryan. The Human Inheritance: Genes, Language, and Evolution (1999)
Cavalli-Sforza, Luigi Luca. "Genes, Peoples, and Languages" (2000)
Sykes, Bryan. "The Seven Daughters of Eve" (2001)
"The Molecule Hunt: Archaeology and the Search for Ancient DNA" by Martin Jones (Archaeological Science, Cambridge Univ.), April 2002.
Copyright © 2001-2010 Kevin F. Duerinck
Animated double helix DNA symbol courtesy of Webpromotion